Prof. Daniel Otzen publishes in Nature Communications

Prof. Daniel Otzen (who recently received the Eliteforsk award) is part of an international research group that recently has uncovered one of the key steps towards fully understanding how spider silk is formed.
They have examined the silk from the funnel-web spider Euprosthenops australis (see picture) on a molecular level.
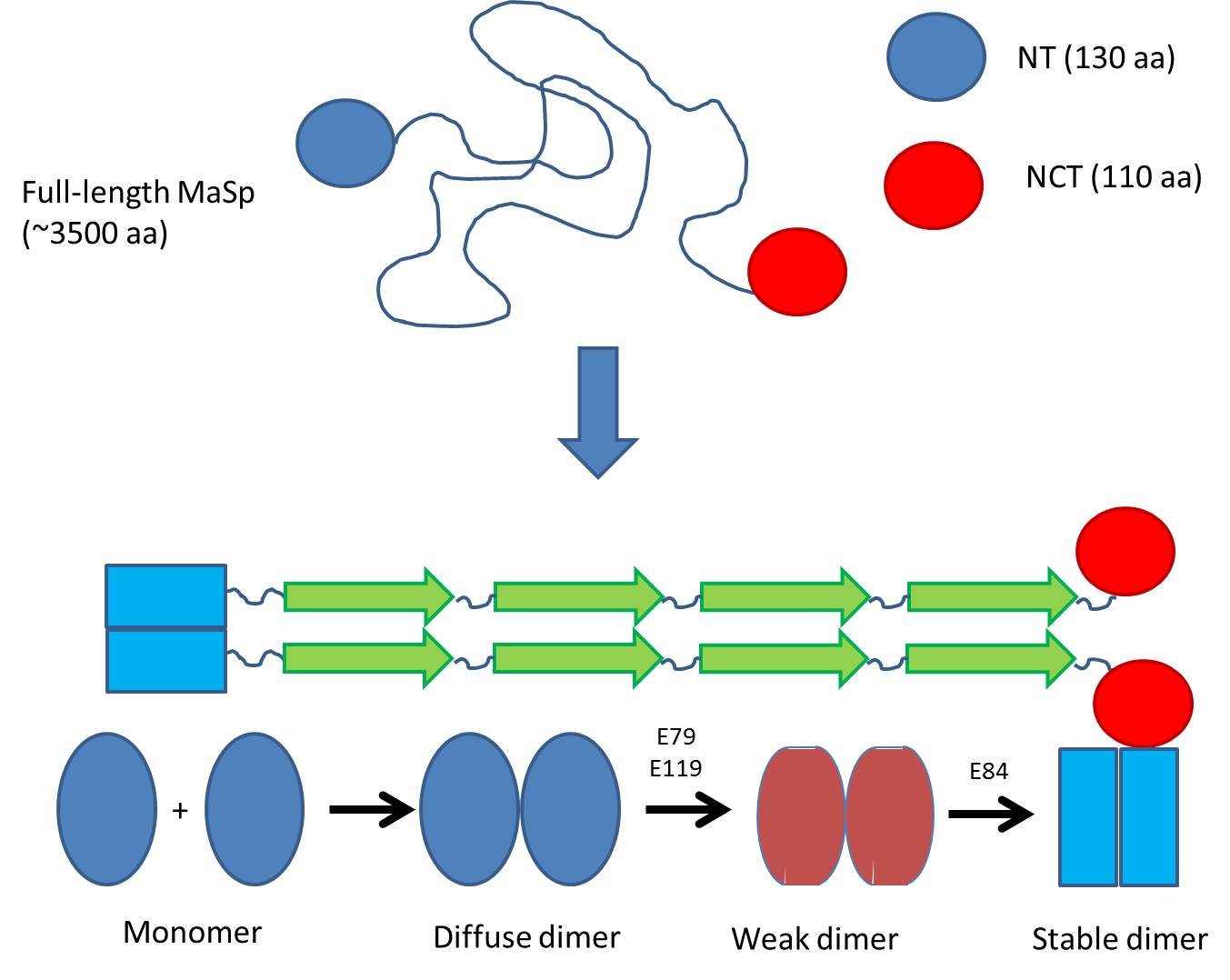
Previously it was known that the silk protein (called MaSp) consists of 3500 amino acids with large regions of polyAla and polyGly, which is stored by spiders at incredibly high concentrations in their silk gland. When excreting the protein, ?-sheet fibrils are formed within seconds as the pH drops from 7 to 5.7.
The research group led by Prof Janne Johansson and Dr. Nina Kronqvist from Karolinska Institutet in Stockholm investigated the N-terminal domain (NT) which is believed to control the fibrillation of the protein. NT is in the monomeric state in the silk gland but forms a dimer as the pH is lowered to around 5.7.
They systematically changed a series of amino acids suspected to be involved in this dimerization and measured how these changes affected the way that the NT monomers dimerize. As pH is gradually lowered from pH 7 in the sac of the silk gland, monomers gradually start to associate weakly, leading to a change in the pKa values of two critical amino acid residues. This allows them to take up protons as early as around pH 6.5, leading to a weak dimer. However, the dimer is only fully stabilized when a final protonation occurs around pH 5.7.
But why is this biologically relevant? Spider silk is formed incredibly rapidly (within seconds); therefore the NT domain, which aligns the different parts of the very large MaSp protein, needs to diffuse and associate very rapidly. The process occurs over a rather broad pH range since the pH drops from around 7 (in the gland where the protein is stored) to around 5.5 when it emerges from the spider. In this way protein is gradually exposed to lower and lower pH, and association can occur in several steps where the first weak associations can be “corrected” if there is wrong assembly. As pH drops to pH 5.5, the dimers are fully stable and silk can be formed. Thus NT dimerization probably helps to align and order the spider silk – an excellent example of biological nanotechnology.
You can find the whole article here
