Breaking news 11th June 2025: We have today in Advanced Materials published the first residue-level resolution structure of a functional bacterial amyloid based solely on experimental data. Read more here.
Our research activities fall within three main topics within protein science.
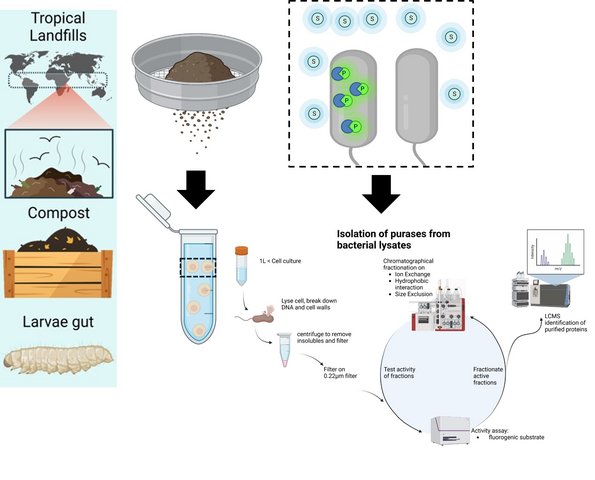
1. Enzymatic plastic degradation. This is mainly carried out within the research center EnZync which involves collaboration between Aarhus University, Danish Technological Institute, the Technical University of Denmark and the University of Porto. This is a multidisciplinary effort coordinated by Daniel Otzen and funded by a Challenge Grant from the Novo Nordisk Foundation. You can read more about EnZync's activities here. We are also funded by the Distinguished Innovator Grant ENCORE to Daniel Otzen.
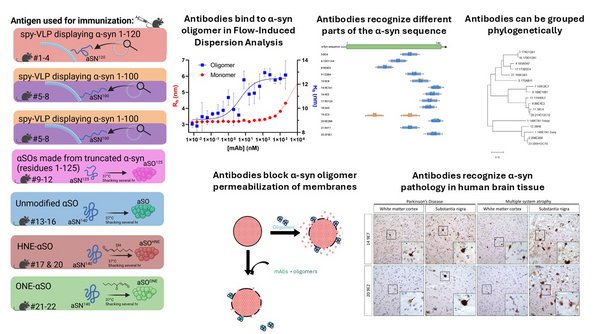
2. Molecular basis of Parkinson's Disease. We explore the mechanisms of aggregation of the protein α-synuclein which is able to form cytotoxic oligomeric and fibrillar species, and devise strategies to combat and contain this aggregation using both antibodies, nanobodies, peptides and small molecules. Our latest efforts focus on the delivery of these therapeutic compounds against α-synuclein aggregation across the blood-brain-barrier using smart nanoliposomes. This takes place within the research center NanoPANS which is funded as a Collaborative Program through the Lundbeck Foundation and is headed by Daniel Otzen. You can read more about the NanoPANS plans and teams here. Work within this area is also financed by a Pioneer Innovator grant PARSOL to Daniel Otzen.
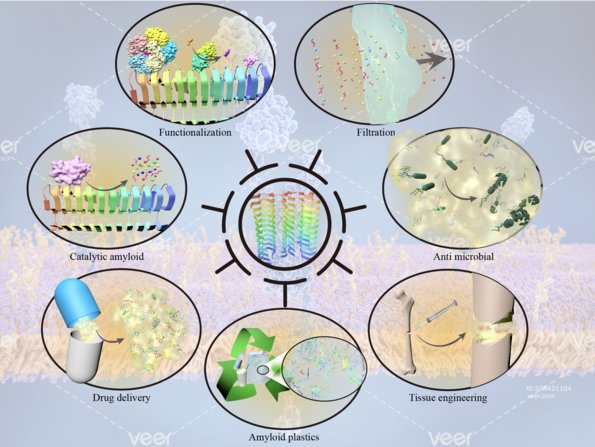
3. Functional amyloid and its uses. We focus on the bacterial amyloid proteins CsgA and FapC, their molecular mechanisms of aggregation and their material properties. This is a close collaboration with Professor Huabing Wang at Guangxi Medical University, Nanning, China. Together with Professor Wang, we have recently solved the structure of FapC and are currently working on strategies to engineer novel uses into them. Overviews of our work can be found here and here.
All our work relates to the study of the kinetics and thermodynamics of protein conformational changes, namely membrane protein folding, protein-detergent interactions and protein fibrillation. These areas are linked by a keen interest in understanding the mechanistic and thermodynamic behaviour of proteins in different circumstances by quantifying the strength of internal side-chain interactions as well as contacts with solvent molecules, whether it be detergents, denaturants, stabilizing salts and osmolytes or lipids. Ultimately we hope this will lead to a greater manipulative ability vis-a-vis processes of both basic, pharmaceutical and industrial relevance. The general approach is to use available spectroscopic techniques (fluorescence, CD, stopped-flow, FTIR, NMR and dynamic and static light scattering) to generate data which can be analyzed in a quantitative manner to develop models and mechanisms for conformational changes at the molecular level.