Article in Nature Chemistry from Harvard's Wyss Institute with iNANO
Designed large DNA crystals could create revolutionary nanodevice
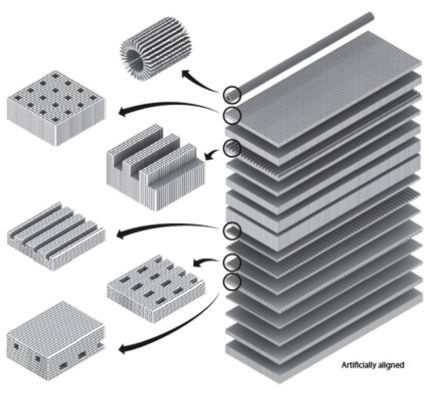
Scientists have long been researching methods for programming behaviours of molecules. Owing largely to its rationalized and justified design strategies, nucleic acids self-assembly, and specifically, DNA self-assembly, this revolutionary method has emerged as a powerful approach in programming self-assembly of custom-shaped intricate one-, two-, and three-dimensional (3D) nanostructures. .
Working with Prof. Yin’s team at Harvard's Wyss Institute for Biologically Inspired Engineering, a novel non-hierarchical crystal growth framework for growing extended crystals in the 3D space was created successfully. Dominant method for DNA crystal growth utilizes a two-stage hierarchal process: individual strands first assemble into a discrete building block (a tile) and individual tiles then assemble into extended lattice structures. Following this strategy, 32 DNA crystals were built with accurately defined depth and an assortment of sophisticated 3D features. Furthermore, the ability to position gold nanoparticles along the crystal structure, inserted with prescribed architectures less than two nanometers apart from each other, was demonstrated.
The work was supported by the Wyss Institute for Biologically Inspired Engineering at Harvard University and the Danish National Research Foundation to the CDNA Centre.
The breakthrough has been published in Nature Chemistry
For further information, please contact:
Postdoc Jie Song song@inano.au.dk
Associate Professor Mingdong Dong dong@inano.au.dk
Centre for DNA Nanotechnology
Interdisciplinary Nanoscience Center, Aarhus University
http://inano.au.dk/biospm/
