Minisymposium
Sensing single RNA and protein conformations in solution - Application of plasmonic Optical Tweezers and nanopores

Info about event
Time
Location
Department of Chemistry, AUD VI Building 1510, Aarhus University
10.15-11.00
Professor Ulrich Felix Keyser, Cavendish Laboratory, University of Cambridge, UK
Title: Nanopore microscope identifies short RNA and RNA isoforms
11.00-11.45
Professor Michael Mayer, Adolphe Merkle Institute, University of Fribourg, CH-1700 Fribourg, Switzerland
Title: Conformational Dynamics of Single Proteins – Exciting Opportunities with Plasmonic
Abstracts:
Professor Ulrich Felix Keyser
Nanopore microscope identifies short RNA and RNA isoforms
Identifying RNA molecules is a remaining challenge in biotechnology. This is driven by the discovery of RNAs that control cellular function ranging in length from a few to 1000s of nucleotides. Here we design three-dimensional nucleic acid constructs that enable the identification of short and long RNA molecules and nanopore readout.
First, we describe the identification of transcript isoforms at the single-molecule level using solid-state nanopore microscopy. We refold target RNA into RNA identifiers with designed sets of complementary DNA strands. Each reshaped molecule carries a unique sequence of structural (pseudo)colours. The sequence of structural colours of RNA identifiers enables simultaneous identification and relative quantification of multiple RNA targets without prior amplification. RNA IDs discriminate circular and linear transcript isoforms in a one-step, enzyme-free reaction in a complex human transcriptome using single-molecule read-out [1].
In the second part, we use designed DNA identifier that allows the multiplexed identification of short RNA molecules. We demonstrate the power of the approach by identifying common viruses and their variants with a nanopores microscope [2].
References:
[1] Nanopore microscope identifies RNA isoforms with structural colours | Nature Chemistry
[2] Simultaneous identification of viruses and their variants with programmable DNA nanobait.
Professor Michael Mayer
Conformational Dynamics of Single Proteins – Exciting Opportunities with Plasmonic Optical Tweezers
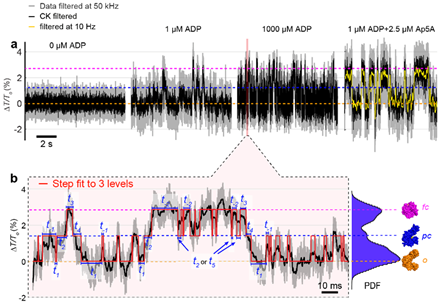
This talk will present an update of our ongoing efforts to assess the potential of nanoplasmonic optical tweezers for interrogating the conformational dynamics of single unmodified proteins in aqueous solution. Specifically, we employ double nanohole (DNH) structures to trap single enzyme proteins [1,2] for minutes to hours. Concurrently, we monitor changes in transmission through the DNH upon exposure of the trapped protein to substrate, product, or inhibitor molecules. We show that experiments with trapped enzymes that are known to undergo significant conformational changes during their catalytic cycle, exhibit multiple, well-defined transmission levels[3]. Increasing concentration of substrate molecules increases the frequency of transitions between these transmission levels in a dose-dependent manner, while the presence of different inhibitors reduces the frequency of transitions by favoring certain transmission levels (Fig. 1). Step-fitting the transmission recordings makes it possible to follow the rate of transition between all levels, revealing individual enzymatic cycles, single molecule turnover frequencies, as well as heretofore unknown enzymatic sub-cycles during catalysis[3]. The talk will conclude with an outlook of applying this approach to additional unmodified enzymes, motor proteins, and transporters as well as a discussion of its current limitations and possible improvements.
References:
[1] Juan, M. L.; Gordon, R.; Pang, Y.; Eftekhari, F.; Quidant, R. 2009, Nat. Phys. 5, 915–919
[2] Pang, Y.; Gordon, R. 2012, Nano Lett. 12, 402–406.
[3] Ying, C.; Karakaci, E.; Bermudez-Urena, E.; Ianiro, A.; Foster, C.; Awasthi, S.; Guha, A.; Bryan, L.; List, J.; Balog, S. 2021, arXiv Prepr, arXiv2107.06407.
[4] Booth, L. S.; Browne, E. V.; Mauranyapin, N. P.; Madsen, L. S.; Barfoot, S.; Mark, A.; Bowen, W. P. 2022, Scientific Reports, 12:1995
