Specialized iNANO Lecture: In silico modeling of biologically complex membranes
Helgi I. Ingólfsson Groningen Biomolecular Sciences and Biotechnology Institute and Zernike Institute for Advanced Materials, University of Groningen, The Netherlands
Info about event
Time
Location
AUD I, Dept. of Chemistry, Langelandsgade 140, 8000 Aarhus C

Helgi I. Ingólfsson, Groningen Biomolecular Sciences and Biotechnology Institute and Zernike Institute for Advanced Materials, University of Groningen, The Netherlands
In silico modeling of biologically complex membranes
The detailed lipid organization of cellular membranes remains elusive. A typical plasma membrane contains hundreds of different lipid species that are actively regulated by the cell. Currently over 40,000 biologically relevant lipids have been identified and specific organisms often synthesize thousands of different lipid types. This is far greater diversity than is needed to maintain bilayer barrier properties and to solvate membrane proteins. Why do organisms go through the costly process of creating and maintaining such a large diversity of lipids? What is the individual role of these lipids, and how do they interact and organize in the membrane plane?
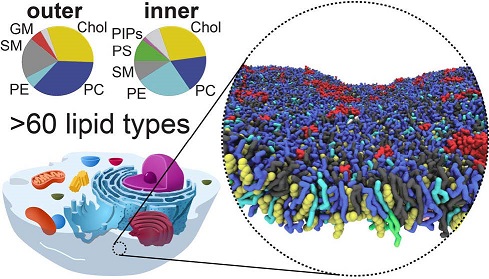
To start to address these questions we model biologically realistic membranes using coarse-grained Martini molecular dynamics simulations. We optimized and developed the Martini lipidome and systematically explored physiochemical properties of >100 different Martini lipid types. Bulk properties of each type (e.g. bilayer thickness, area per lipid, diffusion, order parameter and area compressibility) were analyzed and overall trends compared to experimental values. Biologically realistic idealized membrane compositions were constructed and simulated, such as in (Ingólfsson, et al. Lipid organization of the plasma membrane. JACS, 136:14554-14559, 2014). These large-scale simulations (~70 by 70 nm and multi microsecond long) are in terms of lipid composition by an order of magnitude the most complex simulations to date. They provide a high-resolution view of the lipid organization of biologically relevant membranes; revealing a complex global non-ideal lipid mixing of different species at different spatiotemporal scales. We analyze a variety of membrane physicochemical properties, including: lipid-lipid interactions, bilayer bulk material properties, domain formation and coupling between the bilayer leaflets, for a number of lipid mixtures and conditions.
Hosts: Professor Birgit Schiøtt and postdoc Xavier Periole, Department of Chemistry
