Specialized iNANO Lecture: Crowd Psychology of Nucleic Acid Enzymes
Dr. Matteo Castronovo, Department of Biology, Temple University, Philadelphia, USA & University of Udine, Department of Medical and Biological Sciences, Udine, Italy
Info about event
Time
Location
iNANO Auditorium (1590-012), iNANO House, Gustav Wieds Vej 14, 8000 Aarhus C

Dr. Matteo Castronovo, Department of Biology, Temple University, Philadelphia, USA
&
University of Udine, Department of Medical and Biological Sciences, Udine, Italy
Crowd Psychology of Nucleic Acid Enzymes
Little is known of the effect of confinement on biochemical reactions that occur within nucleic acid nanostructures, in which the type and level of crowding is similar to that of intracellular environments. In this presentation, I will outline novel, unexpected properties of nucleic acid-processing enzymes that arise from nanoscale confinement. Nucleic Acid Density is a Crucial Determinant of Enzyme Diffusion, Recognition, and Reactivity.
In our studies we use AFM nanolithography to form laterally-confined, self-assembled monolayers (“patches’) of short, double-helical nucleic acids on flat gold surfaces, with variable density. We unequivocally show that confinement and density have quantifiable effects on enzyme function.
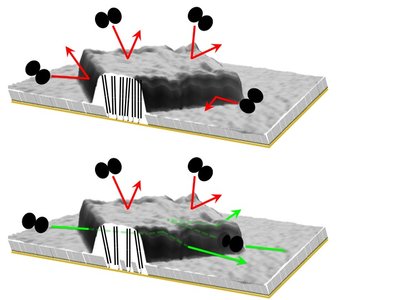 Specifically, for duplex DNA patches we find that (i) restriction endonucleases can gain access to dsDNA matrices with densities lower than a certain critical threshold [1, 2]; (ii) endonucleases can exclusively gain access from the sides, where DNA mobility is reduced, rather than larger, topmost surface, and then can diffuse in a two-dimensional manner inside the matrix [3]; and (iii), if the dsDNA density is higher that a certain critical threshold, and if the monolayer phase is sufficiently homogeneous, the restriction enzyme BamHI can recognize and cleave a non-canonical site with only a partial match with the consensus sequence.
Specifically, for duplex DNA patches we find that (i) restriction endonucleases can gain access to dsDNA matrices with densities lower than a certain critical threshold [1, 2]; (ii) endonucleases can exclusively gain access from the sides, where DNA mobility is reduced, rather than larger, topmost surface, and then can diffuse in a two-dimensional manner inside the matrix [3]; and (iii), if the dsDNA density is higher that a certain critical threshold, and if the monolayer phase is sufficiently homogeneous, the restriction enzyme BamHI can recognize and cleave a non-canonical site with only a partial match with the consensus sequence.
For patches containing hybrid DNA-RNA duplexes, which are substrates for ribonuclease (RNase) H, we find that (iv) an exceptionally large and stable increase in height is observed when the RNase H interacts with the patches in the absence of the catalytic Mg2+ cofactor, indicating the formation of a stable RNase H-hybrid assemblage. (v) In addition, the RNase H-dependent height increase is only observed with RNA-DNA densities above a specific threshold.
These findings demonstrate that nucleic acid processing enzymes may work very differently in crowded nucleic acid nanosystems, than in bulk solution, and that nucleic acid self-assembly offers a method to deliberately control the diffusion, recognition, and reactivity of nucleic acid processing enzymes.
[1] Castronovo, M. et al. Control of Steric Hindrance on Restriction Enzyme Reactions with Surface-Bound DNA Nanostructures. Nano Letters 2008, 8, 4140–4145.
[2] Redhu, S. K.; Castronovo, M. and Nicholson, A.W. Digital Imprinting of RNA Recognition and Processing on a Self-Assembled Nucleic Acid Matrix. Scientific Reports 2013, 3.
[3] Castronovo, M. et al. Two-dimensional Enzyme Diffusion in Laterally Confined DNA Monolayers. Nature Communications 2011, 2, 1–10.
Host: Assistant professor Ebbe Sloth Andersen, Interdisciplinary Nanoscience Center, Aarhus University
