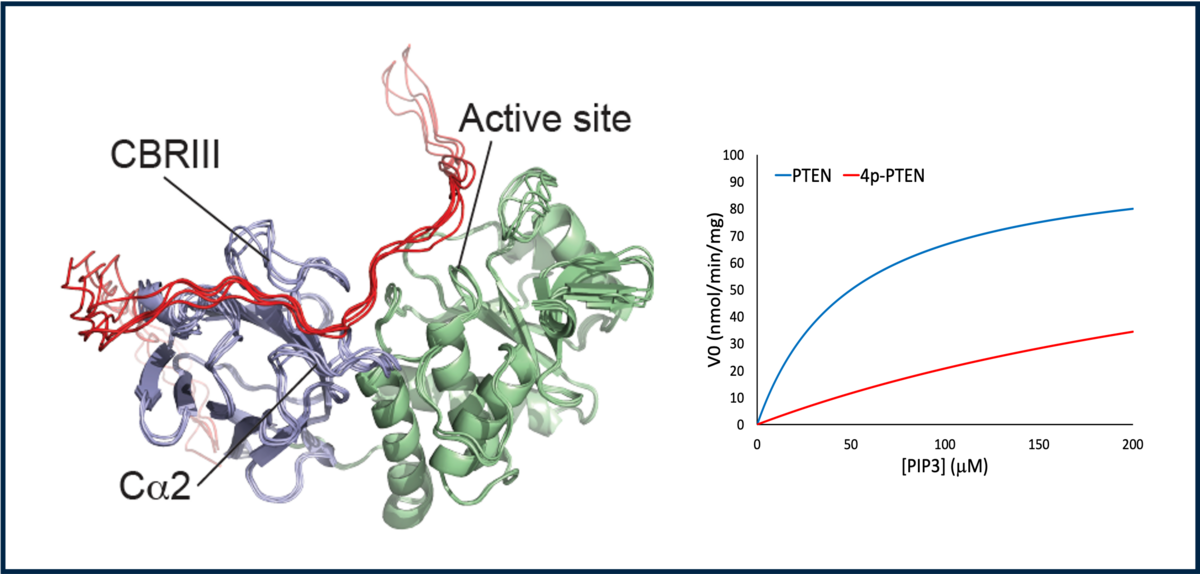
Phosphorylation is a post-translational modification of proteins that is peformed by enzymes called kinases, and erased by phosphatases. Phosphorylation cascades is a way by which cells transfer information from receptors to specific funtions in the cytoplasm or nucleus. Interstingly, this means that kinases are themselves regulated by kinases. These phosphorylation cascades are righly kept in check and their dysregulation often leads to disease. A large body of litterature has characterized kinase pathways and cascades but the molecular determinants of their regulation is still largely elusive.
Our goal is to understand how phosphorylation affects kinases in terms of their conformations, interactions and activity. Phosphorylation modulates kinases by changing the properties of linker regions between structured domains, and by directly participating in auto-inhibition/activation mechanisms.
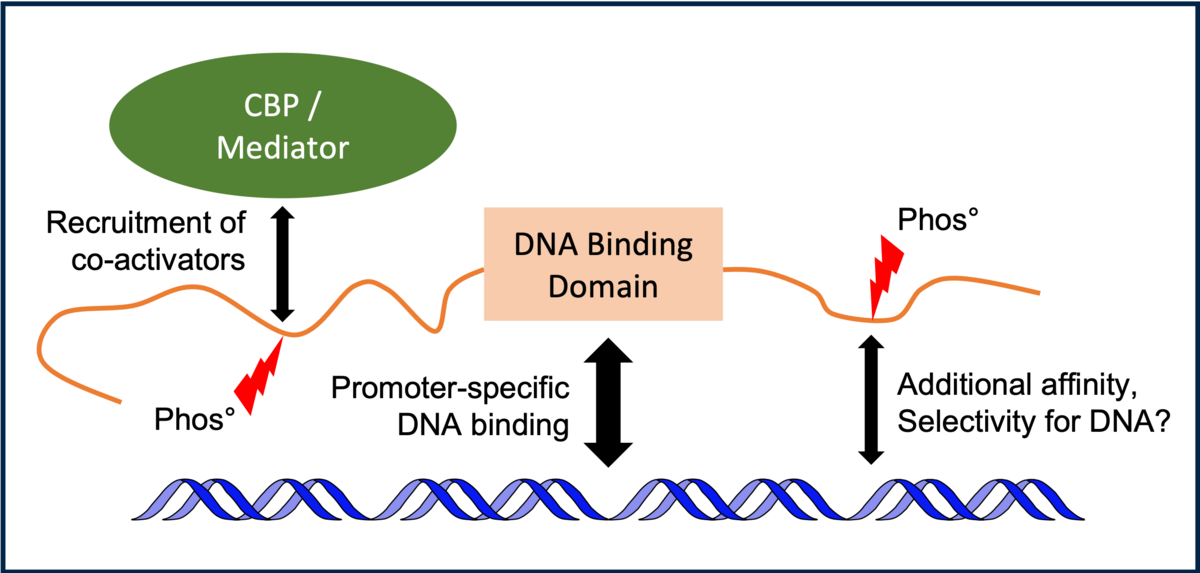
Transcription factors are proteins that regulate the expression of genes (the synthesis of messenger RNA). Post-translational modifications – especially phosphorylation – modulate how transcription factors bind DNA and express a given gene. Phosphorylation sites are found more often in intrinsically disordered regions of transcription factors. Disordered regions and their associated modifications play critical roles in cell signaling, regulation and control which makes them central players in the development of many human diseases.
Our aim is to shed light on the atomistic details of how phosphorylation modulates transcription factor conformation, dynamics and interactions. We strive to understand how this translates into regulation of gene expression at the cellular level in contexts in which cells are dysregulated, e.g. in cancer.
The goal is to develop the best NMR methods tailored for specific biological questions. I have contributed to the development of: