Nanosatellite shows the way to RNA medicine of the future
Researchers from Aarhus University and Berkeley Laboratory have designed RNA molecules, that folds into nanoscale rectangles, cylinders, and satellites, and have studied their 3D structure and dynamics with advanced nanotechnological methods. In an article in the journal Nature Nanotechnology, the researchers describe their work and how it has led to the discovery of rules and mechanisms for RNA folding that will make it possible to build more ideal and functional RNA particles for use in RNA-based medicine.

The RNA molecule is commonly recognized as messenger between DNA and protein, but it can also be folded into intricate molecular machines. An example of a naturally occurring RNA machine is the ribosome, that functions as a protein factory in all cells. Inspired by natural RNA machines, researchers at the Interdisciplinary Nanoscience Center (iNANO) have developed a method called "RNA origami", which makes it possible to design artificial RNA nanostructures that fold from a single stand of RNA. The method is inspired by the Japanese paper folding art, origami, where a single piece of paper can be folded into a given shape, such as a paper bird.
Frozen folds provide new insight
The research paper in Nature Nanotechnology describes how the RNA origami technique was used to design RNA nanostructures, that were characterised by cryo-electron microscopy (cryo-EM) at the Danish National cryo-EM Facility EMBION. Cryo-EM is a method for determining the 3D structure of biomolecules, which works by freezing the sample so quickly that water does not have time to form ice crystals, which means that frozen biomolecules can be observed more clearly with the electron microscope. Images of many thousands of molecules can be converted on the computer into a 3D map, that is used to build an atomic model of the molecule. The cryo-EM investigations provided valuable insight into the detailed structure of the RNA origamis, which allowed optimization of the design process and resulted in more ideal shapes.
“With precise feedback from cryo-EM, we now have the opportunity to fine-tune our molecular designs and construct increasingly intricate nanostructures”, explains Ebbe Sloth Andersen, associate professor at iNANO, Aarhus University.
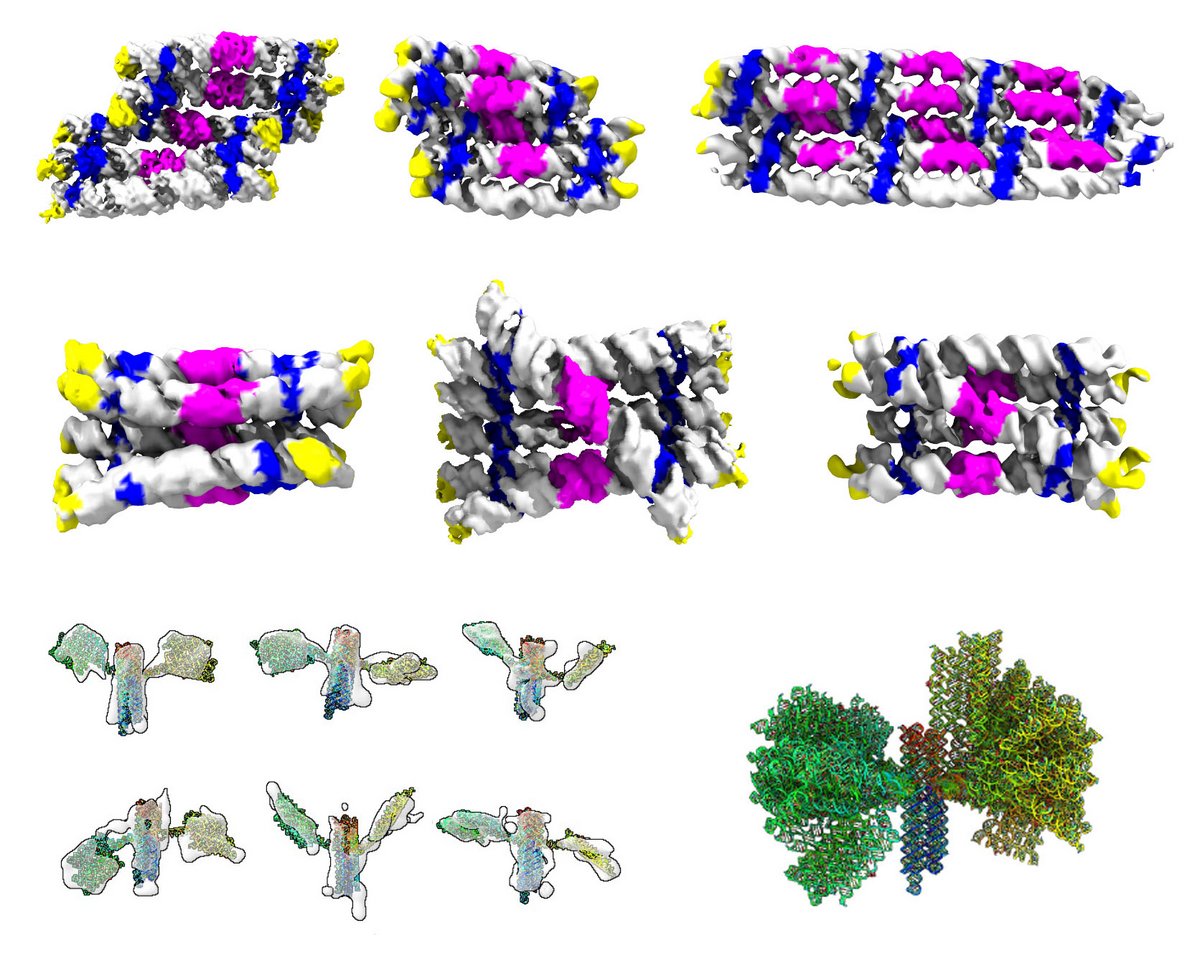
Discovery of a slow folding trap
Cryo-EM images of an RNA cylinder sample turned out to contain two very different shapes, and by freezing the sample at different times it was evident that a transition between the two shapes was taking place. Using the technique of small-angle X-ray scattering (SAXS), where the samples are not frozen, the researchers were able to observe this transition in real time and found that the folding transition occurred after approx. 10 hours. The researchers had discovered a so-called “folding trap” where the RNA gets trapped during transcription and only later gets released(see video).
Video: Video shows the two different shapes of the same RNA molecule and the suggested transition between them. In the first part of the video, a part of the central “kissing loop” interaction is marked in red. In the last part of the video, the base pairs of the central “kissing loop” interaction are shown to highlight how they are broken and reformed during the transition.
“It was quite a surprise to discover an RNA molecule that refolds this slow since folding typically takes place in less than a second” tells Jan Skov Pedersen, Professor at Department of Chemistry and iNANO, Aarhus University.
”We hope to be able to exploit similar mechanisms to activate RNA therapeutics at the right time and place in the patient”, explains Ewan McRae, the first author of the study, who is now starting his own research group at the "Centre for RNA Therapeutics" at the Houston Methodist Research Institute in Texas, USA.
Construction of a nanosatellite from RNA
To demonstrate the formation of complex shapes, the researchers combined RNA rectangles and cylinders to create a multi-domain "nanosatellite" shape, inspired by the Hubble Space Telescope.
"I designed the nanosatellite as a symbol of how RNA design allows us to explore folding space (possibility space of folding) and intracellular space, since the nanosatellite can be expressed in cells", says Cody Geary, assistant professor at iNANO, who originally developed the RNA-origami method.
However, the satellite proved difficult to characterize by cryo-EM due to its flexible properties, so the sample was sent to a laboratory in the USA, where they specialize in determining the 3D structure of individual particles by electron tomography, the so-called IPET-method.
"The RNA satellite was a big challenge! But by using our IPET method, we were able to characterize the 3D shape of individual particles and thus determine the positions of the dynamic solar panels on the nanosatellite", says Gary Ren from the Molecular Foundry at Lawrence Berkeley National Laboratory, California, USA.
The future of RNA medicine
The investigation of the RNA origamis contributes to improving the rational design of RNA molecules for use in medicine and synthetic biology. A new interdisciplinary consortium, COFOLD, supported by the Novo Nordisk Foundation, will continue the investigations of RNA folding processes by involving researchers from computer science, chemistry, molecular biology, and microbiology to design, simulate and measure folding at higher time resolution.
“With the RNA design problem partially solved, the road is now open to creating functional RNA nanostructures that can be used for RNA-based medicine, or act as RNA regulatory elements to reprogram cells”, predicts Ebbe Sloth Andersen.

Related news
Additional information
We strive to ensure that all our articles live up to the Danish universities' principles for good research communication (scroll down to find the English version on the web-site). Because of this the article will be supplemented with the following information: | |
Funding | The studies have received financial support from the Independent Research Fund Denmark, the Natural Sciences and Engineering Research Council of Canada, the European Research Council (ERC), and the Carlsberg Research Foundation. Work at the Molecular Foundry, Berkeley Laboratory, was supported by the Office of Science and Office of Basic Energy Sciences of the U.S. Department of Energy. |
Link to the scientific article | "Structure, folding and flexibility of co-transcriptional RNA origami" Ewan K.S. McRae, Helena Østergaard Rasmussen, Jianfang Liu, Andreas Bøggild, Michael T.A. Nguyen, Nestor Sampedro Vallina, Thomas Boesen, Jan Skov Pedersen, Gang Ren, Cody Geary, Ebbe Sloth Andersen |
Contact | Associate Professor Ebbe Sloth Andersen. Email: esa@inano.au.dk Phone: +45 41178619 |
