Characterization techniques play a key role in the development of nanotechnology. Among various characterization techniques, Atomic Force Microscopy (AFM) is the only one which is widely applied in observing biomolecules and biomaterials down to single molecular resolution in physiological environments. In order to obtain an insight into various biological systems, morphological data is not enough to explain how a myriad of single biomolecules contribute to macro-scale properties and functions. Characterization of the mechanical properties of biological systems at molecular-/nano-scale is one of the desirable ways to reveal more information on how biological systems work as a whole, as it is the intermolecular forces that dominate the communication between individual functional groups, biomolecules and bio-effective units [1, 2]. In other words, mechanical properties are directly related to molecular interactions at the nanoscale, which significantly influence macroscopic functions of tissues and organisms. As a result, nanoscale mechanical analysis is supposed to help trace biological functions and phenomena back to the origin.
As the working principle of AFM is based on interaction force detection, it is considered an attractive solution. AFM-based Force spectroscopy and Force Volume mode have been developed and applied to research for more than a decade. [3] In these modes, the AFM probe works as a force sensor to record force-distance curves during every engagement and retraction cycle. By fitting to a mechanical model, mechanical events are extracted from the force curves as molecular recognition [4] and complex unfolding [5, 6]. However, data acquisition time is too long to follow dynamic interactions and mechanical responses of biomolecules. Therefore, it takes too much time and effort for traditional AFM-based mechanical mapping to reach molecular resolution [7].
Recently, a novel mode of AFM, the so-called quantitative nanomechanical AFM was developed. It narrows the data acquisition interval to microseconds, making it possible to follow dynamic events between biomolecules. Besides, it is capable of providing mechanical maps with nano resolution. More importantly, it is possible to record morphological and mechanical images simultaneously. The relationship between the two images can help determine how a nanoscale feature is related to local mechanical properties[8]. One goal of our group is to explore the applications of quantitative nanomechanical AFM in nanoscale characterization of biological systems and material science.
The followings are several key works in this field.
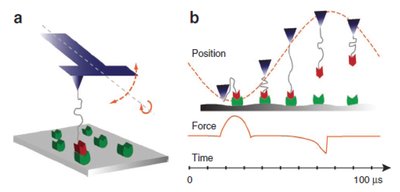
Determination of protein structural flexibility by microsecond force spectroscopy
Proteins are dynamic molecular machines having a structural flexibility that allows conformational changes. Current methods for the determination of protein flexibility rely mainly on the measurement of thermal fluctuations and disorder in protein conformations and tend to be experimentally challenging. Moreover, they reflect atomic fluctuations on picosecond timescales, whereas the large conformational changes in proteins typically happen on micro- to millisecond timescales. Here, we directly determine the flexibility of bacteriorhodopsin—a protein that uses the energy of light to move protons across cell membranes—at the microsecond timescale by monitoring force-induced deformations across the protein structure with a technique based on atomic force microscopy. In contrast to existing methods, the deformations we measure involve a collective response of protein residues and operate under physiologically relevant conditions with native proteins.
Nature Nanotechnology 4, 514 - 517 (2009), doi:10.1038/nnano.2009.156
A nanomechanical interface to rapid single-molecule interactions
Single-molecule techniques provide opportunities for molecularly precise imaging, manipulation, assembly and biophysical studies. Owing to the kinetics of bond rupture processes, rapid single-molecule measurements can reveal novel bond rupture mechanisms, probe single molecule events with short lifetimes and enhance the interaction forces supplied by single molecules. Rapid measurements will also increase throughput necessary for technological use of single-molecule techniques. Here we report a nanomechanical sensor that allows single-molecule force spectroscopy on the previously unexplored microsecond timescale. We probed bond lifetimes around 5 ms and observed significant enhancements in molecular interaction forces. Our loading-rate-dependent measurements provide experimental evidence for an additional energy barrier in the biotin–streptavidin complex. We also demonstrate quantitative mapping of rapid single-molecule interactions with high spatial resolution. This nanomechanical interface may allow studies of molecular processes with short lifetimes and development of novel biological imaging, single-molecule manipulation and assembly technologies.
Nature Communications 2,2011 doi:10.1038/ncomms1246
Quantification of the Interaction Forces between Metals and Graphene
The two-dimensional material graphene has numerous potential applications in nano(opto)electronics, which inevitably involve metal graphene interfaces. Theoretical approaches have been employed to examine metal graphene interfaces, but experimental evidence is currently lacking. Here, we combine atomic force microscopy (AFM) based dynamic force measurements and density functional theory calculations to quantify the interaction between metal-coated AFM tips and graphene under ambient conditions. The results show that copper has the strongest affinity to graphene among the studied metals (Cu, Ag, Au, Pt, Si), which has important implications for the construction of a new generation of electronic devices. Observed differences in the nature of the metal-graphene bonding are well reproduced by the calculations, which included nonlocal Hartree Fock exchange and van der Waals effects.
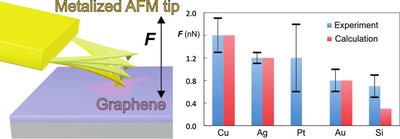
ACS NANO 2013, doi:10.1021/nn305608a.
References
1. Muller, D.J. and Y.F. Dufrene, Atomic force microscopy as a multifunctional molecular toolbox in nanobiotechnology. Nat Nano, 2008. 3(5): p. 261-269.
2. Kurland, N.E., Z. Drira, and V.K. Yadavalli, Measurement of nanomechanical properties of biomolecules using atomic force microscopy. Micron, 2012. 43(2–3): p. 116-128.
3. Müller, D.J. and Y.F. Dufrêne, Force nanoscopy of living cells. Current biology : CB, 2011. 21(6): p. R212-R216.
4. Florin, E., V. Moy, and H. Gaub, Adhesion forces between individual ligand-receptor pairs. Science, 1994. 264(5157): p. 415-417.
5. Rief, M., et al., Single Molecule Force Spectroscopy on Polysaccharides by Atomic Force Microscopy. Science, 1997. 275(5304): p. 1295-1297.
6. Rief, M., et al., Reversible Unfolding of Individual Titin Immunoglobulin Domains by AFM. Science, 1997. 276(5315): p. 1109-1112.
7. Medalsy, I., U. Hensen, and D.J. Muller, Imaging and Quantifying Chemical and Physical Properties of Native Proteins at Molecular Resolution by Force–Volume AFM. Angewandte Chemie International Edition, 2011: p. n/a-n/a.
8. Garcia, R. and E.T. Herruzo, The emergence of multifrequency force microscopy. Nat Nano, 2012. 7(4): p. 217-226.
(For more information: dong@inano.au.dk)
Scanning ion-conductance microscopy (SICM) is a unique scanning probe technique that utilizes a nanopipette for scanning the sample of interest. In the SICM setup, developed by Hansma et al. [1] in 1989, the sample is submersed in an electrolytic solution and the pipette tip is placed in the near vicinity of the sample surface. A potential is applied between an electrode placed inside the pipette and one in the bulk solution, thus creating an ion current through the pipette aperture. The current is mainly determined by the size of the applied potential, the electrolyte conductivity and the aperture size. As the tip approaches the surface of the sample, the ion flow will be occluded. The reduction in current is used to employ a feedback mechanism keeping the tip-sample distance constant.
A key advantage of SICM is that there is no contact between probe and sample, making it ideal for imaging soft materials that might suffer from deformation, when subjected to the force between tip and sample in AFM. We employed SICM in the alternating current mode [2] to investigate the morphology of individual amyloid glucagon fibrils and compared the results to the morphology obtained by liquid and ambient tapping mode AFM. Using SICM (Figure 2A) we obtained results comparable to liquid TM-AFM (E) and superior to ambient TM-AFM (F). We furthermore obtained vertical nanometre resolution, demonstrating the high potential of SICM for a variety of applications.
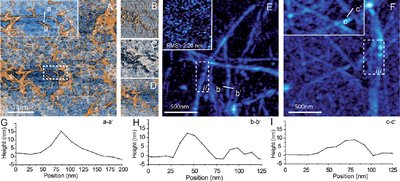
Figure 1: (A) SICM topography image of glucagon fibrils; (B)–(D) the corresponding ICM amplitude, phase and Z detector images; (E) liquid AFM image of glucagon fibrils, the inset image is the Ta surface; (F) air AFM image of glucagon fibrils; (G) the line profile of aa’ in Fig. 2A; (H) the line profile of bb’ in Fig. 2B; and (I) the line profile of cc’ in Fig. 2C.
The ionic current through a nanopipette in a scanning ion-conductance microscopy setup has been observed to be highly dependent on electric charges present on the surface of the glass pipette. This is due to the presence of an electric field and associated electric double layer which leads to a preferential flow of current. This effect is either increased or decreased when the pipette approaches the vicinity of charged surfaces of which nearly all water submersed materials obtain. Surface charge and surface charge density (SCD) are important properties that influence the interaction between surfaces. The uniqueness of SICM allows for nanoscale mapping of SCD under physiological conditions which is not feasible by other methods.
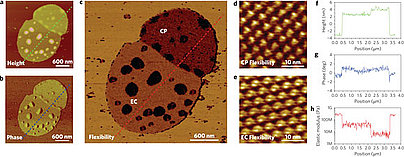
Using this unique capability of SICM, we have obtained important information of biological relevant systems such as DNA, peptide and lipids. Lipids are an integral part of the cellular membrane and lipid SCD determine cell communication, protein regulation and gene delivery. We have studied multiple relevant lipid model systems composed of either single domains [3] or phase-separated multidomains [4] as seen in Figure 2.
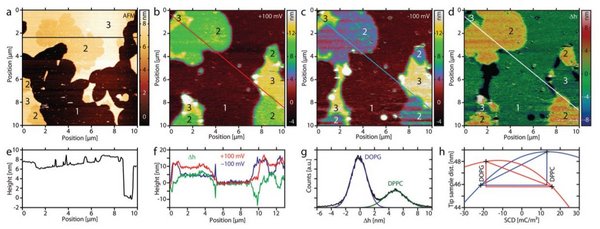
Figure 2: DOPG:DPPC lipids. (a) The AFM image of phase separating domains of DOPG (3) and DPPC (2) on mica (1), the DOPG has formed a double bilayer with height 8 nm and the DPPC is a single bilayer of height 7 nm. The apparent SICM topographies (b), (c) show three domains of distinct height 1: mica support, 2: DPPC (10.1 nm and 4.4 nm), 3: DOPG:DPPC-mix (12.9 nm and 13.2 nm). The CIHD-image (d) calculated from (b) and (c) shows the difference in height of the two apparent topographies, this is proportional to the effective SCD. Panel (f) shows a selected line profile from (b), (c) and (d). The physical height of the domains measured from the AFM image (a), (e) are used to place the relative SCD difference calculated from the histogram (g) on the tip–sample distance vs. the SCD curve (h) to obtain the absolute SCD values.
Our studies of lipid model-systems have shown that apparent lipid height correlate with its surface charge density, which then provide nanoscale SCD information of phase-separated domains in addition to degree of mixture of charged and non-charged lipids. In the future, we aim to perform surface charge mapping on living cells to explore membrane dynamics.
Understanding factors that determine the resolution of SICM measurements
The main advantage of using SICM to measure topography is that it is a non-contact measurement where the ionic conductivity of the electrolyte surrounding the sample is probed. Unlike topography contact techniques, such as atomic force microscopy, SICM enables the study of fragile samples including biological specimens (i.e. cells) without physical perturbation (5). However, SICM does not currently have the resolution capability of AFM. The spatial resolution of SICM measurements is generally considered to be limited by the diameter of the pipette aperture, yet the pipette length and angle are also important parameters (6). When attempting to increase the resolution by implementing nanoscale pipette apertures, the system is further complicated by introducing surface charge effects from the probe and sample that affect ion transport. We are using finite element modelling to further our understanding of how the pipette geometry/material as well as experimental conditions affect the resolution of SICM.
Multifunctional SICM measurements
SICM is widely used to measure topography by utilizing the drop in ionic conductivity occurring between a reference electrode inside the pipette versus another in the electrolyte while the pipette is moved close to a surface. Ionic conductivity is a fundamental parameter arising from the nature of ions present in an electrolyte. This means that SICM can be used for much more than topography measurements as ion transport is involved/changes in scenarios such as crystal dissolution (7), cellular ion transport (8), and in the presence of local surface charges (3). Ions in solution accumulate at charged surfaces and the electric double layer forms. We have previously demonstrated that an effective surface charge can be obtained by measuring the ionic conductivity close to charged surfaces via topography distortions (Figure 1A). We are currently working on using dual-barrel nanopipettes for simultaneous surface charge and topography measurements (Figure 1B). When a potential is applied between two reference electrodes inside each barrel in a dual-barrel pipette, the effects of charge of the pipette is mitigated, restoring Ohmic behavior (Figure C and D), deconvoluting topography, and surface charge responses. Using dual-channel pipettes can further allow us to conduct multifunctional measurements such as ion delivery from the pipette.
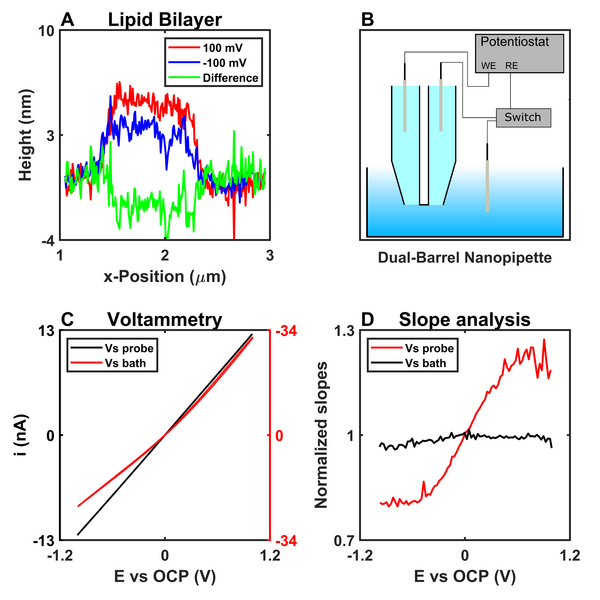
Figure 2: A) Line scans of a DPPG lipid bilayer (Data is reproduced from ref 5). B) Scheme of the dual-barrel method. C) Cyclic voltammetry, showing the mitigation of ion current rectification when applying a potential between the two channels of a dual-barrel pipette. D) Slopes calculated in 41-point intervals of the cyclic voltammograms in C.
[1] Hansma, P. K., Drake, B., Marti, O., Gould, S. A. C., & Prater, C. B. (1989). The scanning ion-conductance microscope. Science, 243(4891), 641-643.
[2] Zhang, Shuai, Sang-Joon Cho, Katerina Busuttil, Chen Wang, Flemming Besenbacher, and Mingdong Dong. "Scanning ion conductance microscopy studies of amyloid fibrils at nanoscale." Nanoscale 4, no. 10 (2012): 3105-3110.
[3] Klausen, L. H., Fuhs, T., & Dong, M. (2016). Mapping surface charge density of lipid bilayers by quantitative surface conductivity microscopy. Nature communications, 7, 12447.
[4] Fuhs, T., Klausen, L. H., Sønderskov, S. M., Han, X., & Dong, M. (2018). Direct measurement of surface charge distribution in phase separating supported lipid bilayers. Nanoscale, 10(9), 4538-4544.
[5] Modern Physics Letters B, 26 (5), 2012, 1130003
[6] Journal of Applied Physics, 105, 2009, 094905
[7] Langmuir, 29 (50), 2013, 15565-155724
[8] Analyst, 37, 2012, 2933-2938
(For more information: lassehyldgaard@inano.au.dk; sts@inano.au.dk; sebska@inano.au.dk)