2D and 3D DNA origami
The DNA origami method allows the folding of long, single-stranded DNA sequences into arbitrary two-dimensional structures by a set of designed oligonucleotides.
Designed iNANO logo origami structures were imaged by high-resolution AFM in liquid. The first and last images are blank 2D origami templates. The middle five ones are the iNANO logo with the modification of biotin- Streptavidin to vary height contrast.

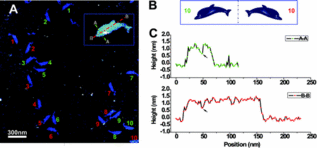
Designed dolphin-like DNA origami structure was imaged by high-resolution AFM in liquid. From the AFM images, it was demonstrated that different designs of the dolphin tail region provided various levels of flexibility in a predictable fashion. Finally, we show that the addition of specific attachment sites promotes dimerization between two independently self-assembled dolphin structures, and that these interactions stabilize the flexible tail.
 z
z
ACS Nano, 2008 2 (6), pp 1213-1218 doi: 10.1021/nn800215j
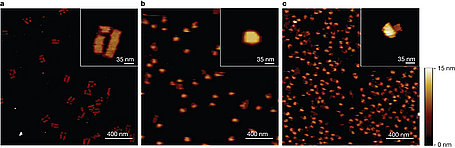
The DNA origami method was extended into three dimensions by creating an addressable DNA box 42 x 36 x 36 nm3 in size that can be opened in the presence of externally supplied DNA 'keys'. We thoroughly characterize the structure of this DNA by AFM.
Nature 459, 73-76 (7 May 2009) doi:10.1038/nature07971
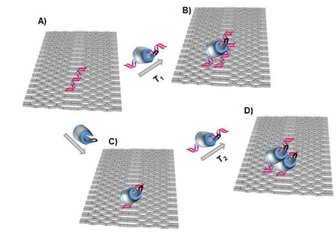
By means of a two-dimensional (2D) DNA origami platform, we monitor the interactions between a single human topoisomerase I covalently bound to one DNA fragment and a second DNA fragment protruding from the DNA origami. This novel single molecule origamibased detection scheme provides direct evidence for the existence of a secondary DNA interaction site in human to poisomerase I and lends further credence to the theory of two distinct DNA interaction sites in human to poisomerase I, possibly facilitating binding to DNA nodes characteristic for plectonemic supercoils.
ACS NANO 4 5969-5977 2010 doi:10.1021/nn101662a
For more information: Jie Song, song@inano.au.dk