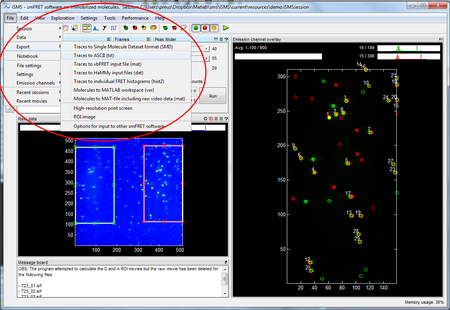
You can export the time series produced using iSMS as pre-formatted input files for other more specialized smFRET software:
For HMM dwell-time analysis you can try out ebFRET, vbFRET, QuB, SMART and/or HaMMy. Note that iSMS already implements HMM analysis by variational Bayesian expectation maximization (the algorithm used in vbFRET). See Quick Guide 3 for more information on how to analyse transitional dynamics using HMM in iSMS.
For thermodynamics and kinetics analysis you can try out SMART and BOBA FRET.
Other export options of the molecule data from iSMS include generic ASCII format, the Single-Molecule Dataset (SMD) format and MATLAB structures.
ebFRET is a successor of vbFRET that analyses multiple FRET traces at once. Ref: Jan-Willem van de Meent, Jonathan E. Bronson, Chris H. Wiggins, Ruben L. Gonzalez Jr. "Empirical Bayes methods enable advanced population-level analyses of single-molecule FRET experiments", Biophys. J. (2014), 106, 6, p1327–1337
SMART is an smFRET analysis toolkit developed in the Herschlag lab at Stanford. Ref: Greenfeld, Max et al. “Single Molecule Analysis Research Tool (SMART): An Integrated Approach for Analyzing Single Molecule Data.” Ed. Daniel J. Muller. PLoS ONE 7.2 (2012): e30024. PMC. Web. 18 Mar. 2015.
Both ebFRET and SMART supports the Single-Molecule Dataset (SMD) format. Ref: Max Greenfeld, Jan-Willem van de Meent, Dmitri S Pavlichin, Hideo Mabuchi, Chris H Wiggins, Ruben L Gonzalez and Daniel Herschlag, "Single-molecule dataset (SMD): a generalized storage format for raw and processed single-molecule data", BMC Bioinformatics (2015), 16:3
First export the data from iSMS to SMD (mat).

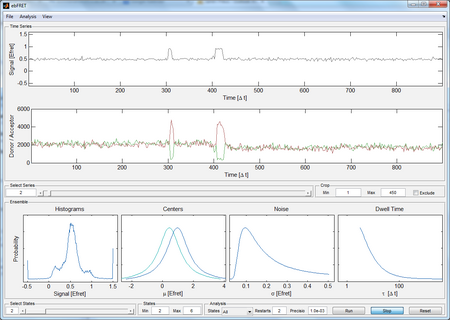
vbFRET is a software that models individual FRET traces using hidden Markov modeling (HMM) and finds the most probable idealized trace using evidence maximization and the variational Bayesian expectation maximization algorithm (VBEM). Reference: Bronson J.E., Fei J., Hofman J.M., Gonzalez R.L., Wiggins C.H. “Learning Rates and States from Biophysical Time Series: A Bayesian Approach to Model Selection and single-molecule FRET Data”. Biophysical Journal 97(12)3196-3205, 2009
HaMMy was the first software specifically designed for HMM analysis of smFRET traces. Ref: S.A. McKinney, C. Joo and T. Ha, "Analysis of single molecule FRET trajectories using hidden Markov modeling", Biophysical Journal (2006)
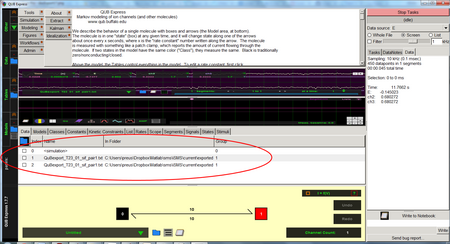
QuB is a software package for HMM time-series analysis originally designed for ion channel experiments. Ref: Milescu, L.S., Nicolai, C., Bannen, J., 2000-2013 QuB Software.
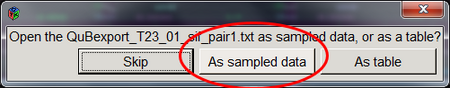
For analysis using QuB export the traces from iSMS as regular ASCII-files (txt).
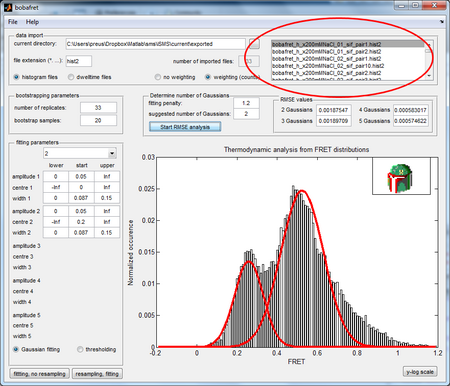
BOBA FRET is a software designed to quantify the cross-sample variability in smFRET data. Ref: Sebastian L. B. König, Mélodie Hadzic, Erica Fiorini, Richard Börner, Danny Kowerko, Wolf U. Blanckenhorn, Roland K. O. Sigel*,BOBA FRET: Bootstrap-based analysis of single-molecule FRET data. PLoS ONE 8(12): e84157
Export molecules from iSMS as hist2 files (single-molecule FRET histograms). When exporting from iSMS you must specify the number of bins and lower and upper bounds for each smFRET histogram (or use the default).